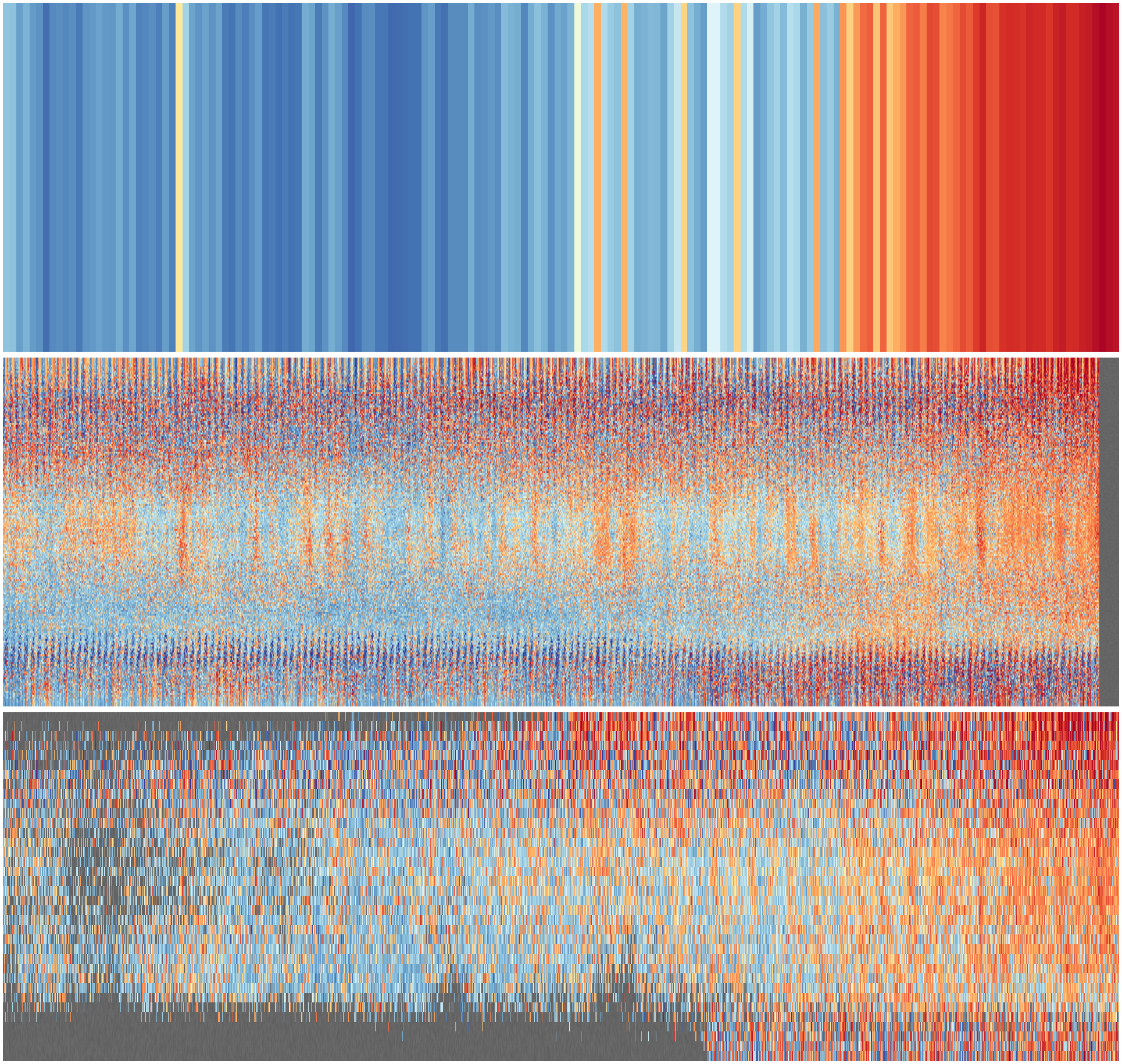
Stripes plotting code¶
Parallise the calculation by extracting the data sample for each year independently:
#!/usr/bin/env python
# Make an latitude slice from Eustace - at a given time,
# sampling across longitude and ensemble.
# Actually, do this for every month in a year - makes a more
# reasonable unit of work
import os
import iris
import numpy
import pickle
import argparse
parser = argparse.ArgumentParser()
parser.add_argument("--year", help="Year",
type=int,required=True)
parser.add_argument("--opdir", help="Directory for output files",
default="%s/EUSTACE/derived/ensemble-monthly" % \
os.getenv('SCRATCH'),
type=str,required=False)
args = parser.parse_args()
if not os.path.isdir(args.opdir):
os.makedirs(args.opdir)
# Fix dask SPICE bug
import dask
dask.config.set(scheduler='single-threaded')
for month in range(1,13):
# Load the Eustace monthly normals
n=iris.load_cube("%s/EUSTACE/1.0/monthly/climatology_1961_1990/%02d.nc" %
(os.getenv('SCRATCH'),month),
iris.Constraint(cube_func=(lambda cell: cell.var_name == 'tas')))
# Array to store the sample in
ndata=numpy.ma.array(numpy.zeros((1,720)),mask=False)
# Load the ensemble (and anomalise)
h=[]
for member in range(10):
e=iris.load_cube("%s/EUSTACE/1.0/monthly/%04d/%02d.nc" %
(os.getenv('SCRATCH'),args.year,month),
iris.Constraint(cube_func=(lambda cell: cell.var_name == 'tasensemble_%d' % member)))
e = e-n # to anomaly
h.append(e)
# Make the slice
for lat in range(720):
member = numpy.random.randint(10)
rand_l = numpy.random.randint(0,1440)
ndata[0,lat]=h[member].data[0,lat,rand_l]
# Store
dfile = "%s/%04d%02d.pkl" % (args.opdir,args.year,month)
pickle.dump( ndata, open( dfile, "wb" ) )
And then running that script for each year as a separate task:
#!/usr/bin/env python
# Scripts to make slices for every month
import os
import datetime
for year in range (1850,2016):
print("./make_slice.py --year=%d" % year )
Then assemble the slices to make the figure:
#!/usr/bin/env python
# Make an extended climate-stripes image from Eustace
# Monthly, resolved in latitude, sampling in longitude,
# sampling across the ensemble.
# Delegates making the slices to the parallelisable make_slice script.
# This script only does the plotting.
import os
import numpy
import datetime
import pickle
import matplotlib
from matplotlib.backends.backend_agg import FigureCanvasAgg as FigureCanvas
from matplotlib.figure import Figure
from matplotlib.patches import Rectangle
from matplotlib.lines import Line2D
start=datetime.datetime(1851,1,1,0,0)
end=datetime.datetime(2018,12,31,23,59)
from get_sample import get_sample_cube
(ndata,dts) = get_sample_cube(start,end)
# Plot the resulting array as a 2d colourmap
fig=Figure(figsize=(19.2,6), # Width, Height (inches)
dpi=300,
facecolor=(0.5,0.5,0.5,1),
edgecolor=None,
linewidth=0.0,
frameon=False,
subplotpars=None,
tight_layout=None)
canvas=FigureCanvas(fig)
matplotlib.rc('image',aspect='auto')
def add_latline(ax,latitude):
latl = (latitude+90)/180
ax.add_line(Line2D([start.timestamp(),end.timestamp()],
[latl,latl],
linewidth=0.5,
color=(0.8,0.8,0.8,1),
zorder=200))
# Add a textured grey background
s=(2000,600)
ax2 = fig.add_axes([0,0.05,1,0.95],facecolor='green')
ax2.set_axis_off() # Don't want surrounding x and y axis
nd2=numpy.random.rand(s[1],s[0])
clrs=[]
for shade in numpy.linspace(.42+.01,.36+.01):
clrs.append((shade,shade,shade,1))
y = numpy.linspace(0,1,s[1])
x = numpy.linspace(0,1,s[0])
img = ax2.pcolormesh(x,y,nd2,
cmap=matplotlib.colors.ListedColormap(clrs),
alpha=1.0,
shading='gouraud',
zorder=10)
# Plot the stripes
ax = fig.add_axes([0,0.05,1,0.95],facecolor='black',
xlim=((start+datetime.timedelta(days=1)).timestamp(),
(end-datetime.timedelta(days=1)).timestamp()),
ylim=(0,1))
ax.set_axis_off()
ndata = numpy.transpose(ndata)
s=ndata.shape
y = numpy.linspace(0,1,s[0]+1)
x = [(a-datetime.timedelta(days=15)).timestamp() for a in dts]
x.append((dts[-1]+datetime.timedelta(days=15)).timestamp())
img = ax.pcolorfast(x,y,numpy.cbrt(ndata),
cmap='RdYlBu_r',
alpha=1.0,
vmin=-1.7,
vmax=1.7,
zorder=100)
for lat in [-60,-30,0,30,60]:
add_latline(ax,lat)
# Add a date grid
axg = fig.add_axes([0,0,1,1],facecolor='green',
xlim=((start+datetime.timedelta(days=1)).timestamp(),
(end-datetime.timedelta(days=1)).timestamp()),
ylim=(0,1))
axg.set_axis_off()
def add_dateline(ax,year):
x = datetime.datetime(year,1,1,0,0).timestamp()
ax.add_line(Line2D([x,x], [0.04,1.0],
linewidth=0.5,
color=(0.8,0.8,0.8,1),
zorder=200))
ax.text(x,0.025,
"%04d" % year,
horizontalalignment='center',
verticalalignment='center',
color='black',
size=14,
clip_on=True,
zorder=200)
for year in range(1860,2020,10):
add_dateline(axg,year)
fig.savefig('ensemble.png')