Default model - validate trained model on test data for a single month¶
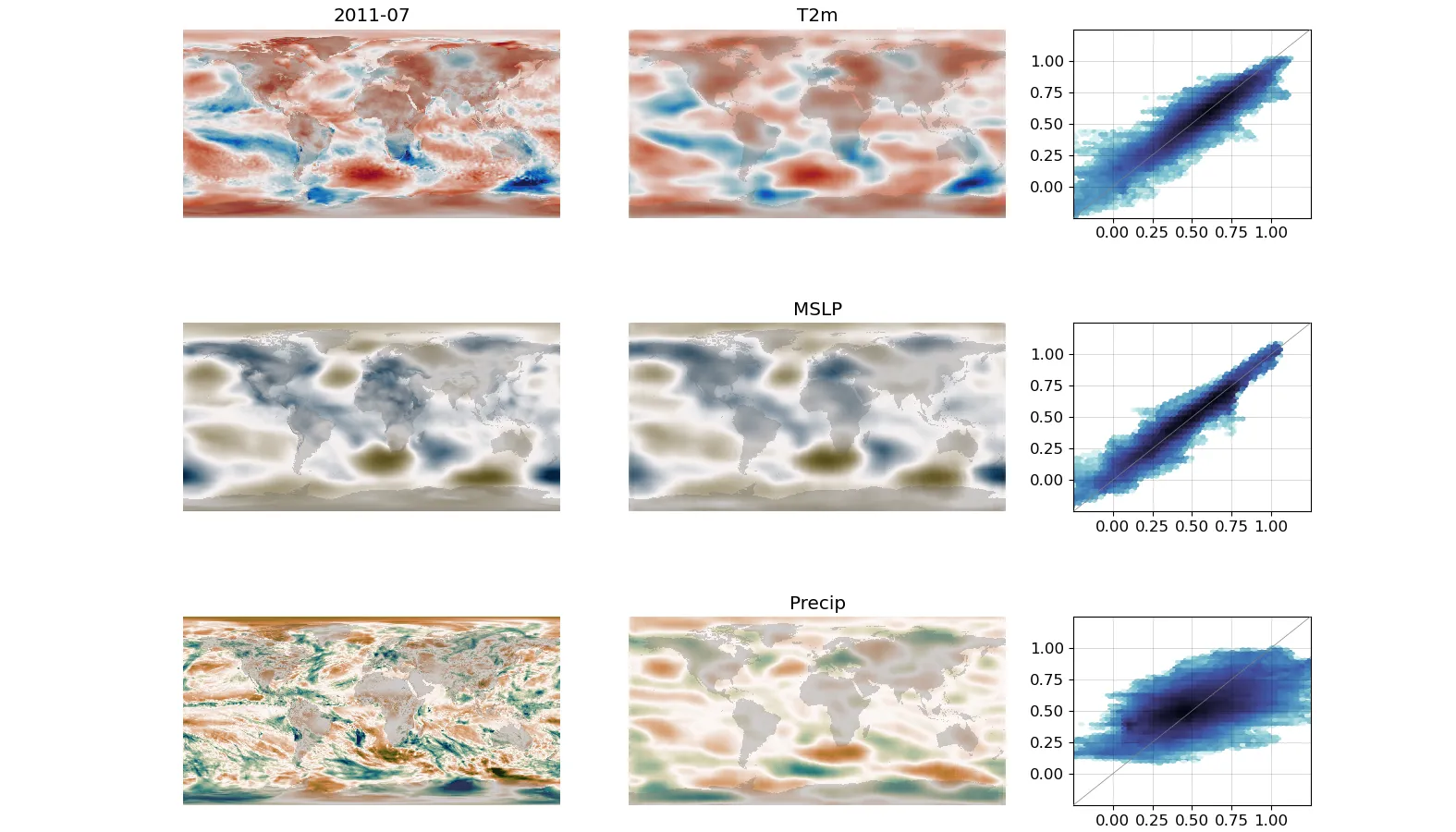
Left-hand column is the target, middle column is the DCVAE output, right-hand column is a scatter plot of target and output.¶
To validate the autoencoder, we can run the trained model using a month from the test dataset as input: The output fields should look like the inputs. Use script validate.py to make the validation figure
By default, it will use a random month from the test set, but you can specify a month using the –year and –month arguments. The –training argument will take months from the training set instead of the test set, and –epoch will use a specific epoch.
#!/usr/bin/env python
# Plot a validation figure for the autoencoder.
# For all outputs:
# 1) Target field
# 2) Autoencoder output
# 3) scatter plot
import os
# Supress TensorFlow moaning about cuda - we don't need a GPU for this
# Also the warning message confuses people.
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
import tensorflow as tf
from ML_models.default.makeDataset import getDataset
from ML_models.default.autoencoderModel import getModel
from ML_models.default.gmUtils import plotValidationField
from specify import specification
strategy = tf.distribute.get_strategy() # No distribution for simple validation
import argparse
parser = argparse.ArgumentParser()
parser.add_argument("--epoch", help="Epoch", type=int, required=False, default=500)
parser.add_argument("--year", help="Test year", type=int, required=False, default=None)
parser.add_argument(
"--month", help="Test month", type=int, required=False, default=None
)
parser.add_argument(
"--training",
help="Use training data (not test)",
default=False,
action="store_true",
)
args = parser.parse_args()
purpose = "Test"
if args.training:
purpose = "Train"
# Go through data and get the desired month
dataset = (
getDataset(specification, purpose=purpose)
.shuffle(specification["shuffleBufferSize"])
.batch(1)
)
input = None
year = None
month = None
for batch in dataset:
dateStr = batch[0][0].numpy().decode("utf-8")
year = int(dateStr[:4])
month = int(dateStr[5:7])
if (args.month is None or month == args.month) and (
args.year is None or year == args.year
):
input = batch
break
if input is None:
raise Exception("Month %04d-%02d not in %s dataset" % (year, month, purpose))
autoencoder = getModel(specification, optimizer=None, epoch=args.epoch)
# Get autoencoded tensors
output = autoencoder.call(input, training=False)
# Make the plot
plotValidationField(specification, input, output, year, month, "comparison.webp")
Utility functions used in the plot
# Model utility functions
import os
import sys
import numpy as np
import datetime
import iris
import tensorflow as tf
from tensorflow.core.util import event_pb2
from tensorflow.python.framework import tensor_util
import matplotlib
from matplotlib.backends.backend_agg import FigureCanvasAgg as FigureCanvas
from matplotlib.figure import Figure
from matplotlib.patches import Rectangle
from matplotlib.lines import Line2D
import cmocean
from utilities import plots, grids
# Convenience function to make everything a list
# lists stay lists, scalars become len=1 lists.
def listify(input):
if isinstance(input, str):
input = [input]
else:
try:
iter(input)
except TypeError:
input = [input]
else:
input = list(input)
return input
# Load the history of a model from the Tensorboard logs
def loadHistory(LSC, offset=-1, max_epoch=None):
history = {}
summary_dir = "%s/DCVAE-Climate/%s/logs/Training" % (os.getenv("SCRATCH"), LSC)
Rfiles = os.listdir(summary_dir)
Rfiles.sort(key=lambda x: os.path.getmtime(os.path.join(summary_dir, x)))
filename = Rfiles[offset]
path = os.path.join(summary_dir, filename)
serialized_records = tf.data.TFRecordDataset(path)
history["epoch"] = []
for srecord in serialized_records:
event = event_pb2.Event.FromString(srecord.numpy())
for value in event.summary.value:
t = tensor_util.MakeNdarray(value.tensor)
if not value.tag in history.keys():
history[value.tag] = []
if value.tag == "OutputNames":
history[value.tag] = t
continue
if len(history[value.tag]) < event.step + 1:
history[value.tag].extend(
[None] * (event.step + 1 - len(history[value.tag]))
)
history[value.tag][event.step] = t
if len(history["epoch"]) < event.step + 1:
history["epoch"].extend([None] * (event.step + 1 - len(history["epoch"])))
history["epoch"][event.step] = event.step
ymax = 0
ymin = 1000000
hts = {}
n_epochs = len(history["Train_loss"])
if max_epoch is not None:
n_epochs = min(max_epoch, n_epochs)
hts["epoch"] = list(range(n_epochs))[1:]
hts["epoch"] = history["epoch"]
for key in history:
if key == "OutputNames":
hts[key] = [str(t, "utf-8") for t in history[key]]
else:
hts[key] = [abs(t) for t in history[key][1:n_epochs] if t is not None]
for key in ("Train_logpz", "Train_logqz_x", "Test_logpz", "Test_logqz_x"):
ymax = max(ymax, max(hts[key]))
ymin = min(ymin, min(hts[key]))
return (hts, ymax, ymin, n_epochs)
# Choose colourmap based on variable name
def get_cmap(name):
if name == "PRATE" or name == "Precip":
return cmocean.cm.tarn
elif name == "MSLP":
return cmocean.cm.diff
else:
return cmocean.cm.balance
# Plot a single-field validation figure for the autoencoder.
def plotValidationField(specification, input, output, year, month, fileName):
nFields = specification["nOutputChannels"]
# Make the plot
figScale = 3.0
wRatios = (2, 2, 1.25)
if specification["trainingMask"] is not None:
wRatios = (2, 2, 1.25, 1.25)
fig = Figure(
figsize=(figScale * sum(wRatios), figScale * nFields),
dpi=100,
facecolor=(1, 1, 1, 1),
edgecolor=None,
linewidth=0.0,
frameon=True,
subplotpars=None,
tight_layout=None,
)
canvas = FigureCanvas(fig)
font = {
"family": "DejaVu Sans",
"sans-serif": "DejaVu Sans",
"weight": "normal",
"size": 12,
}
matplotlib.rc("font", **font)
# Each variable a row in it's own subfigure
subfigs = fig.subfigures(nFields, 1, wspace=0.01)
if nFields == 1:
subfigs = [subfigs]
for varI in range(nFields):
ax_var = subfigs[varI].subplots(
nrows=1, ncols=len(wRatios), width_ratios=wRatios
)
# Left - map of target
varx = grids.E5sCube.copy()
varx.data = np.squeeze(input[-1][:, :, :, varI].numpy())
varx.data = np.ma.masked_where(varx.data == 0.0, varx.data, copy=False)
if varI == 0:
ax_var[0].set_title("%04d-%02d" % (year, month))
ax_var[0].set_axis_off()
x_img = plots.plotFieldAxes(
ax_var[0],
varx,
vMax=1.25,
vMin=-0.25,
cMap=get_cmap(specification["outputNames"][varI]),
)
# Centre - map of model output
vary = grids.E5sCube.copy()
vary.data = np.squeeze(output[:, :, :, varI].numpy())
vary.data = np.ma.masked_where(varx.data == 0.0, vary.data, copy=False)
ax_var[1].set_axis_off()
ax_var[1].set_title(specification["outputNames"][varI])
x_img = plots.plotFieldAxes(
ax_var[1],
vary,
vMax=1.25,
vMin=-0.25,
cMap=get_cmap(specification["outputNames"][varI]),
)
# Third - scatter plot of input::output - where used for training
ax_var[2].set_xticks([0, 0.25, 0.5, 0.75, 1])
ax_var[2].set_yticks([0, 0.25, 0.5, 0.75, 1])
if specification["trainingMask"] is not None:
varxm = varx.copy()
varym = vary.copy()
mflat = specification["trainingMask"].numpy().squeeze()
varxm.data = np.ma.masked_where(mflat == 1, varxm.data, copy=True)
varym.data = np.ma.masked_where(mflat == 1, varym.data, copy=True)
plots.plotScatterAxes(
ax_var[2], varxm, varym, vMin=-0.25, vMax=1.25, bins="log"
)
else:
plots.plotScatterAxes(
ax_var[2], varx, vary, vMin=-0.25, vMax=1.25, bins="log"
)
# Fourth only if masked - scatter plot of input::output - where masked out of training
if specification["trainingMask"] is not None:
ax_var[3].set_xticks([0, 0.25, 0.5, 0.75, 1])
ax_var[3].set_yticks([0, 0.25, 0.5, 0.75, 1])
mflat = specification["trainingMask"].numpy().squeeze()
varx.data = np.ma.masked_where(mflat == 0, varx.data, copy=True)
vary.data = np.ma.masked_where(mflat == 0, vary.data, copy=True)
plots.plotScatterAxes(
ax_var[3], varx, vary, vMin=-0.25, vMax=1.25, bins="log"
)
fig.savefig(fileName)
def plotTrainingMetrics(
specification, hts, fileName="training.webp", chts=None, aymax=None, epoch=None
):
fig = Figure(
figsize=(15, 5),
dpi=100,
facecolor=(1, 1, 1, 1),
edgecolor=None,
linewidth=0.0,
frameon=True,
subplotpars=None,
tight_layout=None,
)
canvas = FigureCanvas(fig)
font = {
"family": "DejaVu Sans",
"sans-serif": "DejaVu Sans",
"weight": "normal",
"size": 12,
}
matplotlib.rc("font", **font)
def addLine(ax, dta, key, col, z, idx=0, rscale=1):
dtp = [listify(x)[idx] for x in dta[key] if len(listify(x)) > idx]
dta2 = [
dta["epoch"][i]
for i in range(len(dta[key]))
if len(listify(dta[key][i])) > idx
]
ax.add_line(
Line2D(
dta2,
np.array(dtp) * rscale,
linewidth=2,
color=col,
alpha=1.0,
zorder=z,
)
)
# Three subfigures
# Left - for the overall loss
# Centre, for the RMS components
# Right, for the KL-divergence components
subfigs = fig.subfigures(1, 3, wspace=0.07)
# Left - Main loss
ymaxL = max(1, max(hts["Train_loss"] + hts["Test_loss"]))
if chts is not None:
ymaxL = max(ymaxL, max(chts["Train_loss"] + chts["Test_loss"]))
if aymax is not None:
ymaxL = aymax
subfigs[0].subplots_adjust(left=0.2)
ax_loss = subfigs[0].subplots(nrows=1, ncols=1)
ax_loss.set_xlim(left=-1, right=epoch + 1, auto=False)
ax_loss.set_ylim(bottom=0, top=ymaxL, auto=False)
ax_loss.set_ylabel("Overall loss")
ax_loss.set_xlabel("epoch")
ax_loss.grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
addLine(ax_loss, hts, "Train_loss", (1, 0.5, 0.5, 1), 10)
addLine(ax_loss, hts, "Test_loss", (1, 0, 0, 1), 20)
if chts is not None:
addLine(ax_loss, chts, "Train_loss", (0.5, 0.5, 1, 1), 10)
addLine(ax_loss, chts, "Test_loss", (0, 0, 1, 1), 20)
# Centre, plot each RMS component as a separate subplot.
comp_font_size = 10
nvar = len(hts["OutputNames"])
# Layout n plots in an (a,b) grid
try:
subplotLayout = [
None,
(1, 1),
(1, 2),
(2, 2),
(2, 2),
(2, 3),
(2, 3),
(3, 3),
(3, 3),
(3, 3),
][nvar + 1]
except Exception:
raise Exception("No subplot layout for %d plots" % nvar)
if subplotLayout is None:
raise Exception("No output names found")
ax_rmse = subfigs[1].subplots(
nrows=subplotLayout[0],
ncols=subplotLayout[1],
sharex=True,
sharey=True,
squeeze=False,
)
ax_rmse = [item for row in ax_rmse for item in row] # Flatten
ax_rmse[0].set_xlim(-1, epoch + 1)
ax_rmse[0].set_ylim(0, ymaxL)
ax_rmse[0].set_ylabel("Variance fraction", fontsize=comp_font_size)
ax_rmse[nvar].set_xlabel("Epoch", fontsize=comp_font_size)
for varI in range(nvar):
ax_rmse[varI].tick_params(axis="both", labelsize=comp_font_size)
ax_rmse[varI].set_title(hts["OutputNames"][varI], fontsize=comp_font_size)
ax_rmse[varI].grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
addLine(ax_rmse[varI], hts, "Train_RMSE", (1, 0.5, 0.5, 1), 10, idx=varI)
addLine(ax_rmse[varI], hts, "Test_RMSE", (1, 0, 0, 1), 20, idx=varI)
if specification["trainingMask"] is not None:
addLine(
ax_rmse[varI], hts, "Train_RMSE_masked", (0.5, 0.5, 1, 1), 10, idx=varI
)
addLine(ax_rmse[varI], hts, "Test_RMSE_masked", (0, 0, 1, 1), 20, idx=varI)
if chts is not None:
for idx in range(len(chts["OutputNames"])):
if chts["OutputNames"][idx] == hts["OutputNames"][varI]:
addLine(
ax_rmse[varI], chts, "Train_RMSE", (0.5, 0.5, 1, 1), 10, idx=idx
)
addLine(ax_rmse[varI], chts, "Test_RMSE", (0, 0, 1, 1), 20, idx=idx)
break
ax_rmse[nvar].tick_params(axis="both", labelsize=comp_font_size)
ax_rmse[nvar].set_title("Regularization", fontsize=comp_font_size)
ax_rmse[nvar].grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
addLine(ax_rmse[nvar], hts, "Regularization_loss", (1, 0, 0, 1), 20)
if chts is not None:
addLine(ax_rmse[nvar], chts, "Regularization_loss", (0, 0, 1, 1), 20)
for varI in range(nvar + 1, len(ax_rmse)):
ax_rmse[varI].set_axis_off()
# Right - KL-divergence plots
ax_kld = subfigs[2].subplots(
nrows=2,
ncols=1,
sharex=True,
sharey=False,
)
ax_kld[0].set_xlim(-1, epoch + 1)
ymaxL = max(hts["Train_logpz"] + hts["Test_logpz"])
yminL = min(hts["Train_logpz"] + hts["Test_logpz"])
if chts is not None:
ymaxL = max(ymaxL, max(chts["Train_logpz"] + chts["Test_logpz"]))
yminL = min(yminL, min(chts["Train_logpz"] + chts["Test_logpz"]))
ymaxL += (ymaxL - yminL) / 20
yminL -= (ymaxL - yminL) / 21
ax_kld[0].set_ylim(yminL, ymaxL)
ax_kld[0].set_title("KL Divergence")
ax_kld[0].set_ylabel("logpz")
ax_kld[0].set_xlabel("")
ax_kld[0].grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
addLine(ax_kld[0], hts, "Train_logpz", (1, 0.5, 0.5, 1), 10)
addLine(ax_kld[0], hts, "Test_logpz", (1, 0, 0, 1), 20)
if chts is not None:
addLine(ax_kld[0], chts, "Train_logpz", (0.5, 0.5, 1, 1), 10)
addLine(ax_kld[0], chts, "Test_logpz", (0, 0, 1, 1), 20)
ymaxL = max(hts["Train_logqz_x"] + hts["Test_logqz_x"])
yminL = min(hts["Train_logqz_x"] + hts["Test_logqz_x"])
if chts is not None:
ymaxL = max(ymaxL, max(chts["Train_logqz_x"] + chts["Test_logqz_x"]))
yminL = min(yminL, min(chts["Train_logqz_x"] + chts["Test_logqz_x"]))
ymaxL += (ymaxL - yminL) / 20
yminL -= (ymaxL - yminL) / 21
ax_kld[1].set_ylim(yminL, ymaxL)
ax_kld[1].set_ylabel("logqz_x")
ax_kld[1].set_xlabel("epoch")
ax_kld[1].grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
addLine(ax_kld[1], hts, "Train_logqz_x", (1, 0.5, 0.5, 1), 10)
addLine(ax_kld[1], hts, "Test_logqz_x", (1, 0, 0, 1), 20)
if chts is not None:
addLine(ax_kld[1], chts, "Train_logqz_x", (0.5, 0.5, 1, 1), 10)
addLine(ax_kld[1], chts, "Test_logqz_x", (0, 0, 1, 1), 20)
# Output as png
fig.savefig(fileName)
# Get target and encoded scalar statistics for one test case
def computeScalarStats(
specification, x, generated, min_lat=-90, max_lat=90, min_lon=-180, max_lon=180
):
nFields = specification["nOutputChannels"]
# get the date from the filename tensor
dateStr = x[0][0].numpy().decode("utf-8")
year = int(dateStr[:4])
month = int(dateStr[5:7])
dtp = datetime.date(year, month, 15)
def tensor_to_cube(t):
result = grids.E5sCube.copy()
result.data = np.squeeze(t.numpy())
result.data = np.ma.masked_where(result.data == 0.0, result.data, copy=False)
return result
def field_to_scalar(field):
field = field.extract(
iris.Constraint(
coord_values={"grid_latitude": lambda cell: min_lat <= cell <= max_lat}
)
& iris.Constraint(
coord_values={"grid_longitude": lambda cell: min_lon <= cell <= max_lon}
)
)
return np.mean(field.data)
stats = {}
stats["dtp"] = dtp
stats["target"] = {}
stats["generated"] = {}
for varI in range(nFields):
if specification["trainingMask"] is None:
stats["target"][specification["outputNames"][varI]] = field_to_scalar(
tensor_to_cube(tf.squeeze(x[-1][:, :, :, varI])),
)
stats["generated"][specification["outputNames"][varI]] = field_to_scalar(
tensor_to_cube(tf.squeeze(generated[:, :, :, varI])),
)
else:
mask = specification["trainingMask"].numpy().squeeze()
stats["target"][specification["outputNames"][varI]] = field_to_scalar(
tensor_to_cube(tf.squeeze(x[-1][:, :, :, varI] * mask)),
)
stats["target"]["%s_masked" % specification["outputNames"][varI]] = (
field_to_scalar(
tensor_to_cube(tf.squeeze(x[-1][:, :, :, varI] * (1 - mask))),
)
)
stats["generated"][specification["outputNames"][varI]] = field_to_scalar(
tensor_to_cube(tf.squeeze(generated[:, :, :, varI] * mask)),
)
stats["generated"]["%s_masked" % specification["outputNames"][varI]] = (
field_to_scalar(
tensor_to_cube(tf.squeeze(generated[:, :, :, varI] * (1 - mask))),
)
)
return stats
def plotScalarStats(all_stats, specification, fileName="multi.webp"):
nFields = specification["nOutputChannels"]
if specification["trainingMask"] is not None:
nFields *= 2
figScale = 3.0
wRatios = (3, 1.25)
# Make the plot
fig = Figure(
figsize=(figScale * sum(wRatios), figScale * nFields),
dpi=300,
facecolor=(1, 1, 1, 1),
edgecolor=None,
linewidth=0.0,
frameon=True,
subplotpars=None,
tight_layout=None,
)
canvas = FigureCanvas(fig)
font = {
"family": "DejaVu Sans",
"sans-serif": "DejaVu Sans",
"weight": "normal",
"size": 14,
}
matplotlib.rc("font", **font)
# Plot a variable in its subfigure
def plot_var(sfig, ts, t, m, label):
# Get two axes in the subfig
var_axes = sfig.subplots(nrows=1, ncols=2, width_ratios=wRatios, squeeze=False)
# Calculate y range
ymin = min(min(t), min(m))
ymax = max(max(t), max(m))
ypad = (ymax - ymin) * 0.1
if ypad == 0:
ypad = 1
# First subaxis for time-series plot
var_axes[0, 0].set_xlim(
ts[0] - datetime.timedelta(days=15), ts[-1] + datetime.timedelta(days=15)
)
var_axes[0, 0].set_ylim(ymin - ypad, ymax + ypad)
var_axes[0, 0].grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
var_axes[0, 0].text(
ts[0] - datetime.timedelta(days=15),
ymax + ypad,
label,
ha="left",
va="top",
bbox=dict(boxstyle="square,pad=0.5", fc=(1, 1, 1, 1)),
zorder=100,
)
var_axes[0, 0].add_line(
Line2D(ts, t, linewidth=2, color=(0, 0, 0, 1), alpha=1.0, zorder=50)
)
var_axes[0, 0].add_line(
Line2D(ts, m, linewidth=2, color=(1, 0, 0, 1), alpha=1.0, zorder=60)
)
# Second subaxis for scatter plot
var_axes[0, 1].set_xlim(ymin - ypad, ymax + ypad),
var_axes[0, 1].set_ylim(ymin - ypad, ymax + ypad),
var_axes[0, 1].grid(color=(0, 0, 0, 1), linestyle="-", linewidth=0.1)
var_axes[0, 1].scatter(t, m, s=2, color=(1, 0, 0, 1), zorder=60)
var_axes[0, 1].add_line(
Line2D(
(ymin - ypad, ymax + ypad),
(ymin - ypad, ymax + ypad),
linewidth=1,
color=(0, 0, 0, 1),
alpha=0.2,
zorder=10,
)
)
# Each variable in its own subfig
subfigs = fig.subfigures(nFields, 1, wspace=0.01)
if nFields == 1:
subfigs = [subfigs]
for varI in range(len(specification["outputNames"])):
if specification["trainingMask"] is None:
vName = specification["outputNames"][varI]
plot_var(
subfigs[varI],
all_stats["dtp"],
all_stats["target"][vName],
all_stats["generated"][vName],
vName,
)
else:
vName = specification["outputNames"][varI]
plot_var(
subfigs[varI * 2],
all_stats["dtp"],
all_stats["target"][vName],
all_stats["generated"][vName],
vName,
)
vName = "%s_masked" % specification["outputNames"][varI]
plot_var(
subfigs[varI * 2 + 1],
all_stats["dtp"],
all_stats["target"][vName],
all_stats["generated"][vName],
specification["outputNames"][varI] + " (masked)",
)
fig.savefig(fileName)
