Estimate normalization parameters for ERA5 data¶
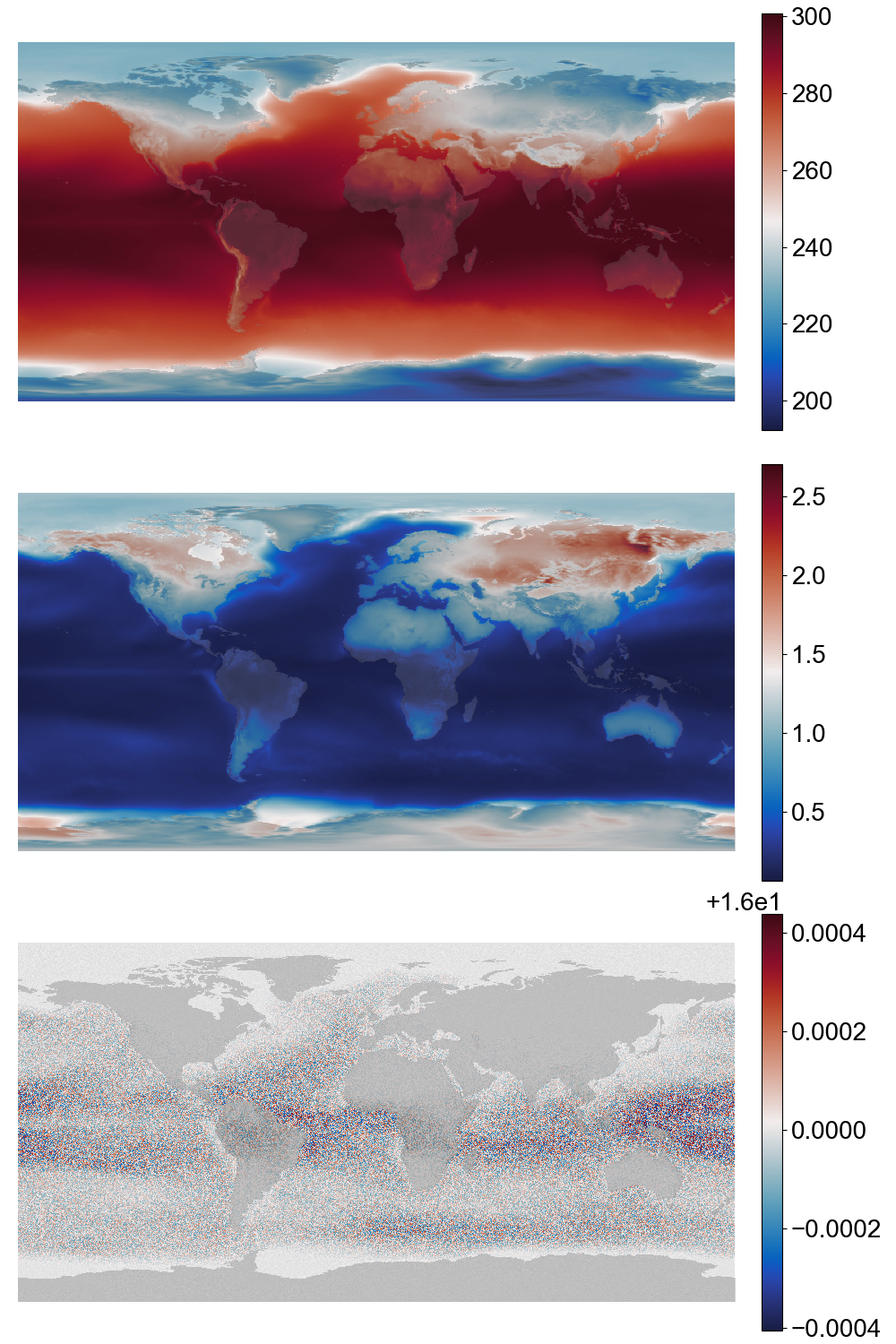
Fitted gamma distribution parameters for temperature data. Top: location, centre: shape, bottom: scale (details.¶
To normalize the data we are fitting gamma distributions to the training data. We fit a separate distribution at every grid-point, for each calendar month (so 1 distribution for each point, for all January data, and so on).
The scripts to estimate the fits are in the normalize directory. The script normalize_all.sh creates a set of commands to estimate all the fits. The script outputs a list of other scripts (one per year, month, variable). Running all the output scripts will do the estimation. (Use GNU parallel to run the scripts efficiently - or submit them as jobs to a cluster).
#!/bin/bash
# Make normalization constants for all the datasets
# Requires pre-made raw tensors
(cd ERA5 && ./make_all_fits.py)
Other scripts used by that main script:
Script to make normalization parameters for one calendar month. Takes arguments –month, -variable, –startyear, and –endyear:
#!/usr/bin/env python
# Find optimum gamma parameters by fitting to the data moments
import os
import sys
import iris
from utilities import grids
import numpy as np
# Supress iris moaning about attributes
iris.FUTURE.save_split_attrs = True
# Supress TensorFlow moaning about cuda - we don't need a GPU for this
# Also the warning message confuses people.
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
import tensorflow as tf
from makeDataset import getDataset
import argparse
parser = argparse.ArgumentParser()
parser.add_argument("--month", help="Month to fit", type=int, required=True)
parser.add_argument("--variable", help="Variable", type=str, required=True)
parser.add_argument(
"--startyear", help="Start Year", type=int, required=False, default=1850
)
parser.add_argument(
"--endyear", help="End Year", type=int, required=False, default=2050
)
parser.add_argument(
"--opdir",
help="Directory for output files",
default="%s/DCVAE-Climate/normalization/ERA5" % os.getenv("SCRATCH"),
)
args = parser.parse_args()
opdir = "%s/%s" % (args.opdir, args.variable)
if not os.path.isdir(opdir):
os.makedirs(opdir, exist_ok=True)
# Go through data and estimate min and mean (for each month)
trainingData = getDataset(
args.variable,
startyear=args.startyear,
endyear=args.endyear,
cache=False,
blur=None,
).batch(1)
mean = tf.zeros([1, 721, 1440, 1], dtype=tf.float32)
count = tf.zeros([1, 721, 1440, 1], dtype=tf.float32)
cnt = 0
for batch in trainingData:
month = int(batch[1].numpy()[0][5:7])
if month == args.month:
valid = ~tf.math.is_nan(batch[0])
mean = tf.where(valid, mean + batch[0] * 3, mean)
count = tf.where(valid, count + 3.0, count)
if (
month == args.month + 1
or month == args.month - 1
or (args.month == 1 and month == 12)
or (args.month == 12 and month == 1)
):
valid = ~tf.math.is_nan(batch[0])
mean = tf.where(valid, mean + batch[0], mean)
count = tf.where(valid, count + 1.0, count)
enough = count >= 10
mean = tf.where(enough, mean / count, np.nan)
count *= 0
# Go through again, now we know means, and estimate variance
variance = tf.zeros([721, 1440, 1], dtype=tf.float32)
for batch in trainingData:
month = int(batch[1].numpy()[0][5:7])
if month == args.month:
valid = tf.math.logical_and(~tf.math.is_nan(batch[0]), ~tf.math.is_nan(mean))
variance = tf.where(
valid, variance + tf.math.squared_difference(batch[0], mean) * 3, variance
)
count = tf.where(valid, count + 3.0, count)
if (
month == args.month + 1
or month == args.month - 1
or (args.month == 1 and month == 12)
or (args.month == 12 and month == 1)
):
valid = tf.math.logical_and(~tf.math.is_nan(batch[0]), ~tf.math.is_nan(mean))
variance = tf.where(
valid, variance + tf.math.squared_difference(batch[0], mean), variance
)
count = tf.where(valid, count + 1.0, count)
enough = count >= 10
variance = tf.where(enough, variance / count, np.nan)
# Artificially expand under-sea-ice variability in SST
if args.variable == "sea_surface_temperature":
variance = tf.where(np.logical_and(enough, mean < 273), variance + 0.5, variance)
# Gamma parameter estimates:
fg_location = tf.zeros([1, 721, 1440, 1], dtype=tf.float32)
fg_location = tf.where(enough, mean - tf.sqrt(variance) * 4, np.nan)
mean = tf.where(enough, mean - fg_location, np.nan)
fg_scale = tf.where(enough, variance / mean, np.nan)
fg_shape = tf.where(enough, mean / fg_scale, np.nan)
shape = grids.E5sCube.copy()
shape.data = np.squeeze(fg_shape.numpy())
shape.data = np.ma.MaskedArray(shape.data, np.isnan(shape.data))
shape.data.data[shape.data.mask] = 1.0
iris.save(
shape,
"%s/%s/shape_m%02d.nc" % (args.opdir, args.variable, args.month),
)
location = grids.E5sCube.copy()
location.data = np.squeeze(fg_location.numpy())
location.data = np.ma.MaskedArray(location.data, np.isnan(location.data))
location.data.data[location.data.mask] = -1.0
iris.save(
location,
"%s/%s/location_m%02d.nc" % (args.opdir, args.variable, args.month),
)
scale = grids.E5sCube.copy()
scale.data = np.squeeze(fg_scale.numpy())
scale.data = np.ma.MaskedArray(scale.data, np.isnan(scale.data))
scale.data.data[scale.data.mask] = 1.0
iris.save(
scale,
"%s/%s/scale_m%02d.nc" % (args.opdir, args.variable, args.month),
)
The data are taken from the tf.tensor datasets of raw data created during the normalization process. Functions to present these as tf.data.DataSets:
# Create raw data dataset for normalization
import os
import sys
import tensorflow as tf
import numpy as np
import zarr
import tensorstore as ts
# Get a dataset - all the tensors for a given and variable
def getDataset(
variable,
startyear=None,
endyear=None,
blur=None,
cache=False,
):
# Get the index of the last month in the raw tensors
fn = "%s/DCVAE-Climate/raw_datasets/ERA5/%s_zarr" % (
os.getenv("SCRATCH"),
variable,
)
zarr_array = zarr.open(fn, mode="r")
AvailableMonths = zarr_array.attrs["AvailableMonths"]
dates = sorted(AvailableMonths.keys())
indices = [AvailableMonths[date] for date in dates]
# Create TensorFlow Dataset object from the source file dates
tn_data = tf.data.Dataset.from_tensor_slices(tf.constant(dates, tf.string))
ts_data = tf.data.Dataset.from_tensor_slices(tf.constant(indices, tf.int32))
# Convert from list ofavailable months to Dataset of source file contents
tsa = ts.open(
{
"driver": "zarr",
"kvstore": "file://" + fn,
}
).result()
# Need the indirect function as zarr can't take tensor indices and .map prohibits .numpy()
def load_tensor_from_index_py(idx):
return tf.convert_to_tensor(tsa[:, :, idx.numpy()].read().result(), tf.float32)
def load_tensor_from_index(idx):
result = tf.py_function(
load_tensor_from_index_py,
[idx],
tf.float32,
)
result = tf.reshape(result, [721, 1440, 1])
return result
ts_data = ts_data.map(
load_tensor_from_index, num_parallel_calls=tf.data.experimental.AUTOTUNE
)
# Add noise to data - needed for some cases where the data is all zero
if blur is not None:
ts_data = ts_data.map(
lambda x: x + tf.random.normal([721, 1440, 1], stddev=blur),
num_parallel_calls=tf.data.experimental.AUTOTUNE,
)
# Zip the data together with the years (so we can find the date and source of each
# data tensor if we need it).
tz_data = tf.data.Dataset.zip((ts_data, tn_data))
# Optimisation
if cache:
tz_data = tz_data.cache() # Great, iff you have enough RAM for it
tz_data = tz_data.prefetch(tf.data.experimental.AUTOTUNE)
return tz_data
